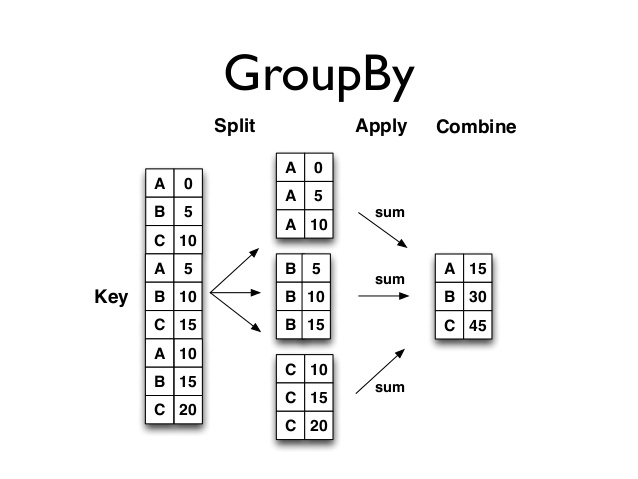
In this quick notes, we will go through aggregation in Python.
This is part of lectures on Learning Python for Data Analysis and Visualization by Jose Portilla on Udemy.
import numpy as np
import pandas as pd
from pandas import Series, DataFrame
url = “http://archive.ics.uci.edu/ml/machine-learning-databases/wine-quality/”
In [2]:
dframe_wine = pd.read_csv('winequality-red.csv', sep=';')
In [4]:
dframe_wine.head()
Out[4]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
| 1 | 7.8 | 0.88 | 0.00 | 2.6 | 0.098 | 25.0 | 67.0 | 0.9968 | 3.20 | 0.68 | 9.8 | 5 |
| 2 | 7.8 | 0.76 | 0.04 | 2.3 | 0.092 | 15.0 | 54.0 | 0.9970 | 3.26 | 0.65 | 9.8 | 5 |
| 3 | 11.2 | 0.28 | 0.56 | 1.9 | 0.075 | 17.0 | 60.0 | 0.9980 | 3.16 | 0.58 | 9.8 | 6 |
| 4 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | 5 |
In [5]:
#get the average alcohol content for all the wines
dframe_wine['alcohol'].mean()
Out[5]:
10.422983114446502
In [6]:
#define a function which will return the differnce between the min and maximum values
def max_to_min(arr):
return arr.max() - arr.min()
In [7]:
wino = dframe_wine.groupby('quality')
wino.describe()
Out[7]:
| alcohol | chlorides | citric acid | density | fixed acidity | free sulfur dioxide | pH | residual sugar | sulphates | total sulfur dioxide | volatile acidity | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | ||||||||||||
| 3 | count | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 | 10.000000 |
| mean | 9.955000 | 0.122500 | 0.171000 | 0.997464 | 8.360000 | 11.000000 | 3.398000 | 2.635000 | 0.570000 | 24.900000 | 0.884500 | |
| std | 0.818009 | 0.066241 | 0.250664 | 0.002002 | 1.770875 | 9.763879 | 0.144052 | 1.401596 | 0.122020 | 16.828877 | 0.331256 | |
| min | 8.400000 | 0.061000 | 0.000000 | 0.994710 | 6.700000 | 3.000000 | 3.160000 | 1.200000 | 0.400000 | 9.000000 | 0.440000 | |
| 25% | 9.725000 | 0.079000 | 0.005000 | 0.996150 | 7.150000 | 5.000000 | 3.312500 | 1.875000 | 0.512500 | 12.500000 | 0.647500 | |
| 50% | 9.925000 | 0.090500 | 0.035000 | 0.997565 | 7.500000 | 6.000000 | 3.390000 | 2.100000 | 0.545000 | 15.000000 | 0.845000 | |
| 75% | 10.575000 | 0.143000 | 0.327500 | 0.998770 | 9.875000 | 14.500000 | 3.495000 | 3.100000 | 0.615000 | 42.500000 | 1.010000 | |
| max | 11.000000 | 0.267000 | 0.660000 | 1.000800 | 11.600000 | 34.000000 | 3.630000 | 5.700000 | 0.860000 | 49.000000 | 1.580000 | |
| 4 | count | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 | 53.000000 |
| mean | 10.265094 | 0.090679 | 0.174151 | 0.996542 | 7.779245 | 12.264151 | 3.381509 | 2.694340 | 0.596415 | 36.245283 | 0.693962 | |
| std | 0.934776 | 0.076192 | 0.201030 | 0.001575 | 1.626624 | 9.025926 | 0.181441 | 1.789436 | 0.239391 | 27.583374 | 0.220110 | |
| min | 9.000000 | 0.045000 | 0.000000 | 0.993400 | 4.600000 | 3.000000 | 2.740000 | 1.300000 | 0.330000 | 7.000000 | 0.230000 | |
| 25% | 9.600000 | 0.067000 | 0.030000 | 0.995650 | 6.800000 | 6.000000 | 3.300000 | 1.900000 | 0.490000 | 14.000000 | 0.530000 | |
| 50% | 10.000000 | 0.080000 | 0.090000 | 0.996500 | 7.500000 | 11.000000 | 3.370000 | 2.100000 | 0.560000 | 26.000000 | 0.670000 | |
| 75% | 11.000000 | 0.089000 | 0.270000 | 0.997450 | 8.400000 | 15.000000 | 3.500000 | 2.800000 | 0.600000 | 49.000000 | 0.870000 | |
| max | 13.100000 | 0.610000 | 1.000000 | 1.001000 | 12.500000 | 41.000000 | 3.900000 | 12.900000 | 2.000000 | 119.000000 | 1.130000 | |
| 5 | count | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 | 681.000000 |
| mean | 9.899706 | 0.092736 | 0.243686 | 0.997104 | 8.167254 | 16.983847 | 3.304949 | 2.528855 | 0.620969 | 56.513950 | 0.577041 | |
| std | 0.736521 | 0.053707 | 0.180003 | 0.001589 | 1.563988 | 10.955446 | 0.150618 | 1.359753 | 0.171062 | 36.993116 | 0.164801 | |
| min | 8.500000 | 0.039000 | 0.000000 | 0.992560 | 5.000000 | 3.000000 | 2.880000 | 1.200000 | 0.370000 | 6.000000 | 0.180000 | |
| 25% | 9.400000 | 0.074000 | 0.090000 | 0.996200 | 7.100000 | 9.000000 | 3.200000 | 1.900000 | 0.530000 | 26.000000 | 0.460000 | |
| 50% | 9.700000 | 0.081000 | 0.230000 | 0.997000 | 7.800000 | 15.000000 | 3.300000 | 2.200000 | 0.580000 | 47.000000 | 0.580000 | |
| 75% | 10.200000 | 0.094000 | 0.360000 | 0.997900 | 8.900000 | 23.000000 | 3.400000 | 2.600000 | 0.660000 | 84.000000 | 0.670000 | |
| max | 14.900000 | 0.611000 | 0.790000 | 1.003150 | 15.900000 | 68.000000 | 3.740000 | 15.500000 | 1.980000 | 155.000000 | 1.330000 | |
| 6 | count | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 | 638.000000 |
| mean | 10.629519 | 0.084956 | 0.273824 | 0.996615 | 8.347179 | 15.711599 | 3.318072 | 2.477194 | 0.675329 | 40.869906 | 0.497484 | |
| std | 1.049639 | 0.039563 | 0.195108 | 0.002000 | 1.797849 | 9.940911 | 0.153995 | 1.441576 | 0.158650 | 25.038250 | 0.160962 | |
| min | 8.400000 | 0.034000 | 0.000000 | 0.990070 | 4.700000 | 1.000000 | 2.860000 | 0.900000 | 0.400000 | 6.000000 | 0.160000 | |
| 25% | 9.800000 | 0.068250 | 0.090000 | 0.995402 | 7.000000 | 8.000000 | 3.220000 | 1.900000 | 0.580000 | 23.000000 | 0.380000 | |
| 50% | 10.500000 | 0.078000 | 0.260000 | 0.996560 | 7.900000 | 14.000000 | 3.320000 | 2.200000 | 0.640000 | 35.000000 | 0.490000 | |
| 75% | 11.300000 | 0.088000 | 0.430000 | 0.997893 | 9.400000 | 21.000000 | 3.410000 | 2.500000 | 0.750000 | 54.000000 | 0.600000 | |
| max | 14.000000 | 0.415000 | 0.780000 | 1.003690 | 14.300000 | 72.000000 | 4.010000 | 15.400000 | 1.950000 | 165.000000 | 1.040000 | |
| 7 | count | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 | 199.000000 |
| mean | 11.465913 | 0.076588 | 0.375176 | 0.996104 | 8.872362 | 14.045226 | 3.290754 | 2.720603 | 0.741256 | 35.020101 | 0.403920 | |
| std | 0.961933 | 0.029456 | 0.194432 | 0.002176 | 1.992483 | 10.175255 | 0.150101 | 1.371509 | 0.135639 | 33.191206 | 0.145224 | |
| min | 9.200000 | 0.012000 | 0.000000 | 0.990640 | 4.900000 | 3.000000 | 2.920000 | 1.200000 | 0.390000 | 7.000000 | 0.120000 | |
| 25% | 10.800000 | 0.062000 | 0.305000 | 0.994765 | 7.400000 | 6.000000 | 3.200000 | 2.000000 | 0.650000 | 17.500000 | 0.300000 | |
| 50% | 11.500000 | 0.073000 | 0.400000 | 0.995770 | 8.800000 | 11.000000 | 3.280000 | 2.300000 | 0.740000 | 27.000000 | 0.370000 | |
| 75% | 12.100000 | 0.087000 | 0.490000 | 0.997360 | 10.100000 | 18.000000 | 3.380000 | 2.750000 | 0.830000 | 43.000000 | 0.485000 | |
| max | 14.000000 | 0.358000 | 0.760000 | 1.003200 | 15.600000 | 54.000000 | 3.780000 | 8.900000 | 1.360000 | 289.000000 | 0.915000 | |
| 8 | count | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 | 18.000000 |
| mean | 12.094444 | 0.068444 | 0.391111 | 0.995212 | 8.566667 | 13.277778 | 3.267222 | 2.577778 | 0.767778 | 33.444444 | 0.423333 | |
| std | 1.224011 | 0.011678 | 0.199526 | 0.002378 | 2.119656 | 11.155613 | 0.200640 | 1.295038 | 0.115379 | 25.433240 | 0.144914 | |
| min | 9.800000 | 0.044000 | 0.030000 | 0.990800 | 5.000000 | 3.000000 | 2.880000 | 1.400000 | 0.630000 | 12.000000 | 0.260000 | |
| 25% | 11.325000 | 0.062000 | 0.302500 | 0.994175 | 7.250000 | 6.000000 | 3.162500 | 1.800000 | 0.690000 | 16.000000 | 0.335000 | |
| 50% | 12.150000 | 0.070500 | 0.420000 | 0.994940 | 8.250000 | 7.500000 | 3.230000 | 2.100000 | 0.740000 | 21.500000 | 0.370000 | |
| 75% | 12.875000 | 0.075500 | 0.530000 | 0.997200 | 10.225000 | 16.500000 | 3.350000 | 2.600000 | 0.820000 | 43.000000 | 0.472500 | |
| max | 14.000000 | 0.086000 | 0.720000 | 0.998800 | 12.600000 | 42.000000 | 3.720000 | 6.400000 | 1.100000 | 88.000000 | 0.850000 |
In [8]:
#do an aggregation on the groupby object
wino.agg(max_to_min)
Out[8]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | |||||||||||
| 3 | 4.9 | 1.140 | 0.66 | 4.5 | 0.206 | 31.0 | 40.0 | 0.00609 | 0.47 | 0.46 | 2.6 |
| 4 | 7.9 | 0.900 | 1.00 | 11.6 | 0.565 | 38.0 | 112.0 | 0.00760 | 1.16 | 1.67 | 4.1 |
| 5 | 10.9 | 1.150 | 0.79 | 14.3 | 0.572 | 65.0 | 149.0 | 0.01059 | 0.86 | 1.61 | 6.4 |
| 6 | 9.6 | 0.880 | 0.78 | 14.5 | 0.381 | 71.0 | 159.0 | 0.01362 | 1.15 | 1.55 | 5.6 |
| 7 | 10.7 | 0.795 | 0.76 | 7.7 | 0.346 | 51.0 | 282.0 | 0.01256 | 0.86 | 0.97 | 4.8 |
| 8 | 7.6 | 0.590 | 0.69 | 5.0 | 0.042 | 39.0 | 76.0 | 0.00800 | 0.84 | 0.47 | 4.2 |
In [16]:
wino.agg(sum)
Out[16]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | |||||||||||
| 3 | 83.6 | 8.845 | 1.71 | 26.35 | 1.225 | 110.0 | 249.0 | 9.97464 | 33.98 | 5.70 | 99.550000 |
| 4 | 412.3 | 36.780 | 9.23 | 142.80 | 4.806 | 650.0 | 1921.0 | 52.81675 | 179.22 | 31.61 | 544.050000 |
| 5 | 5561.9 | 392.965 | 165.95 | 1722.15 | 63.153 | 11566.0 | 38486.0 | 679.02757 | 2250.67 | 422.88 | 6741.700000 |
| 6 | 5325.5 | 317.395 | 174.70 | 1580.45 | 54.202 | 10024.0 | 26075.0 | 635.84041 | 2116.93 | 430.86 | 6781.633333 |
| 7 | 1765.6 | 80.380 | 74.66 | 541.40 | 15.241 | 2795.0 | 6969.0 | 198.22475 | 654.86 | 147.51 | 2281.716667 |
| 8 | 154.2 | 7.620 | 7.04 | 46.40 | 1.232 | 239.0 | 602.0 | 17.91382 | 58.81 | 13.82 | 217.700000 |
In [14]:
wino.agg('count')
Out[14]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | |||||||||||
| 3 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| 4 | 53 | 53 | 53 | 53 | 53 | 53 | 53 | 53 | 53 | 53 | 53 |
| 5 | 681 | 681 | 681 | 681 | 681 | 681 | 681 | 681 | 681 | 681 | 681 |
| 6 | 638 | 638 | 638 | 638 | 638 | 638 | 638 | 638 | 638 | 638 | 638 |
| 7 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 199 | 199 |
| 8 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 | 18 |
In [17]:
#create a new column in dataframe as per your requirements
dframe_wine['alc / quality ratio'] = dframe_wine['alcohol'] / dframe_wine['quality']
In [18]:
dframe_wine
Out[18]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | alc / quality ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.4 | 0.700 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.99780 | 3.51 | 0.56 | 9.4 | 5 | 1.880000 |
| 1 | 7.8 | 0.880 | 0.00 | 2.6 | 0.098 | 25.0 | 67.0 | 0.99680 | 3.20 | 0.68 | 9.8 | 5 | 1.960000 |
| 2 | 7.8 | 0.760 | 0.04 | 2.3 | 0.092 | 15.0 | 54.0 | 0.99700 | 3.26 | 0.65 | 9.8 | 5 | 1.960000 |
| 3 | 11.2 | 0.280 | 0.56 | 1.9 | 0.075 | 17.0 | 60.0 | 0.99800 | 3.16 | 0.58 | 9.8 | 6 | 1.633333 |
| 4 | 7.4 | 0.700 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.99780 | 3.51 | 0.56 | 9.4 | 5 | 1.880000 |
| 5 | 7.4 | 0.660 | 0.00 | 1.8 | 0.075 | 13.0 | 40.0 | 0.99780 | 3.51 | 0.56 | 9.4 | 5 | 1.880000 |
| 6 | 7.9 | 0.600 | 0.06 | 1.6 | 0.069 | 15.0 | 59.0 | 0.99640 | 3.30 | 0.46 | 9.4 | 5 | 1.880000 |
| 7 | 7.3 | 0.650 | 0.00 | 1.2 | 0.065 | 15.0 | 21.0 | 0.99460 | 3.39 | 0.47 | 10.0 | 7 | 1.428571 |
| 8 | 7.8 | 0.580 | 0.02 | 2.0 | 0.073 | 9.0 | 18.0 | 0.99680 | 3.36 | 0.57 | 9.5 | 7 | 1.357143 |
| 9 | 7.5 | 0.500 | 0.36 | 6.1 | 0.071 | 17.0 | 102.0 | 0.99780 | 3.35 | 0.80 | 10.5 | 5 | 2.100000 |
| 10 | 6.7 | 0.580 | 0.08 | 1.8 | 0.097 | 15.0 | 65.0 | 0.99590 | 3.28 | 0.54 | 9.2 | 5 | 1.840000 |
| 11 | 7.5 | 0.500 | 0.36 | 6.1 | 0.071 | 17.0 | 102.0 | 0.99780 | 3.35 | 0.80 | 10.5 | 5 | 2.100000 |
| 12 | 5.6 | 0.615 | 0.00 | 1.6 | 0.089 | 16.0 | 59.0 | 0.99430 | 3.58 | 0.52 | 9.9 | 5 | 1.980000 |
| 13 | 7.8 | 0.610 | 0.29 | 1.6 | 0.114 | 9.0 | 29.0 | 0.99740 | 3.26 | 1.56 | 9.1 | 5 | 1.820000 |
| 14 | 8.9 | 0.620 | 0.18 | 3.8 | 0.176 | 52.0 | 145.0 | 0.99860 | 3.16 | 0.88 | 9.2 | 5 | 1.840000 |
| 15 | 8.9 | 0.620 | 0.19 | 3.9 | 0.170 | 51.0 | 148.0 | 0.99860 | 3.17 | 0.93 | 9.2 | 5 | 1.840000 |
| 16 | 8.5 | 0.280 | 0.56 | 1.8 | 0.092 | 35.0 | 103.0 | 0.99690 | 3.30 | 0.75 | 10.5 | 7 | 1.500000 |
| 17 | 8.1 | 0.560 | 0.28 | 1.7 | 0.368 | 16.0 | 56.0 | 0.99680 | 3.11 | 1.28 | 9.3 | 5 | 1.860000 |
| 18 | 7.4 | 0.590 | 0.08 | 4.4 | 0.086 | 6.0 | 29.0 | 0.99740 | 3.38 | 0.50 | 9.0 | 4 | 2.250000 |
| 19 | 7.9 | 0.320 | 0.51 | 1.8 | 0.341 | 17.0 | 56.0 | 0.99690 | 3.04 | 1.08 | 9.2 | 6 | 1.533333 |
| 20 | 8.9 | 0.220 | 0.48 | 1.8 | 0.077 | 29.0 | 60.0 | 0.99680 | 3.39 | 0.53 | 9.4 | 6 | 1.566667 |
| 21 | 7.6 | 0.390 | 0.31 | 2.3 | 0.082 | 23.0 | 71.0 | 0.99820 | 3.52 | 0.65 | 9.7 | 5 | 1.940000 |
| 22 | 7.9 | 0.430 | 0.21 | 1.6 | 0.106 | 10.0 | 37.0 | 0.99660 | 3.17 | 0.91 | 9.5 | 5 | 1.900000 |
| 23 | 8.5 | 0.490 | 0.11 | 2.3 | 0.084 | 9.0 | 67.0 | 0.99680 | 3.17 | 0.53 | 9.4 | 5 | 1.880000 |
| 24 | 6.9 | 0.400 | 0.14 | 2.4 | 0.085 | 21.0 | 40.0 | 0.99680 | 3.43 | 0.63 | 9.7 | 6 | 1.616667 |
| 25 | 6.3 | 0.390 | 0.16 | 1.4 | 0.080 | 11.0 | 23.0 | 0.99550 | 3.34 | 0.56 | 9.3 | 5 | 1.860000 |
| 26 | 7.6 | 0.410 | 0.24 | 1.8 | 0.080 | 4.0 | 11.0 | 0.99620 | 3.28 | 0.59 | 9.5 | 5 | 1.900000 |
| 27 | 7.9 | 0.430 | 0.21 | 1.6 | 0.106 | 10.0 | 37.0 | 0.99660 | 3.17 | 0.91 | 9.5 | 5 | 1.900000 |
| 28 | 7.1 | 0.710 | 0.00 | 1.9 | 0.080 | 14.0 | 35.0 | 0.99720 | 3.47 | 0.55 | 9.4 | 5 | 1.880000 |
| 29 | 7.8 | 0.645 | 0.00 | 2.0 | 0.082 | 8.0 | 16.0 | 0.99640 | 3.38 | 0.59 | 9.8 | 6 | 1.633333 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 1569 | 6.2 | 0.510 | 0.14 | 1.9 | 0.056 | 15.0 | 34.0 | 0.99396 | 3.48 | 0.57 | 11.5 | 6 | 1.916667 |
| 1570 | 6.4 | 0.360 | 0.53 | 2.2 | 0.230 | 19.0 | 35.0 | 0.99340 | 3.37 | 0.93 | 12.4 | 6 | 2.066667 |
| 1571 | 6.4 | 0.380 | 0.14 | 2.2 | 0.038 | 15.0 | 25.0 | 0.99514 | 3.44 | 0.65 | 11.1 | 6 | 1.850000 |
| 1572 | 7.3 | 0.690 | 0.32 | 2.2 | 0.069 | 35.0 | 104.0 | 0.99632 | 3.33 | 0.51 | 9.5 | 5 | 1.900000 |
| 1573 | 6.0 | 0.580 | 0.20 | 2.4 | 0.075 | 15.0 | 50.0 | 0.99467 | 3.58 | 0.67 | 12.5 | 6 | 2.083333 |
| 1574 | 5.6 | 0.310 | 0.78 | 13.9 | 0.074 | 23.0 | 92.0 | 0.99677 | 3.39 | 0.48 | 10.5 | 6 | 1.750000 |
| 1575 | 7.5 | 0.520 | 0.40 | 2.2 | 0.060 | 12.0 | 20.0 | 0.99474 | 3.26 | 0.64 | 11.8 | 6 | 1.966667 |
| 1576 | 8.0 | 0.300 | 0.63 | 1.6 | 0.081 | 16.0 | 29.0 | 0.99588 | 3.30 | 0.78 | 10.8 | 6 | 1.800000 |
| 1577 | 6.2 | 0.700 | 0.15 | 5.1 | 0.076 | 13.0 | 27.0 | 0.99622 | 3.54 | 0.60 | 11.9 | 6 | 1.983333 |
| 1578 | 6.8 | 0.670 | 0.15 | 1.8 | 0.118 | 13.0 | 20.0 | 0.99540 | 3.42 | 0.67 | 11.3 | 6 | 1.883333 |
| 1579 | 6.2 | 0.560 | 0.09 | 1.7 | 0.053 | 24.0 | 32.0 | 0.99402 | 3.54 | 0.60 | 11.3 | 5 | 2.260000 |
| 1580 | 7.4 | 0.350 | 0.33 | 2.4 | 0.068 | 9.0 | 26.0 | 0.99470 | 3.36 | 0.60 | 11.9 | 6 | 1.983333 |
| 1581 | 6.2 | 0.560 | 0.09 | 1.7 | 0.053 | 24.0 | 32.0 | 0.99402 | 3.54 | 0.60 | 11.3 | 5 | 2.260000 |
| 1582 | 6.1 | 0.715 | 0.10 | 2.6 | 0.053 | 13.0 | 27.0 | 0.99362 | 3.57 | 0.50 | 11.9 | 5 | 2.380000 |
| 1583 | 6.2 | 0.460 | 0.29 | 2.1 | 0.074 | 32.0 | 98.0 | 0.99578 | 3.33 | 0.62 | 9.8 | 5 | 1.960000 |
| 1584 | 6.7 | 0.320 | 0.44 | 2.4 | 0.061 | 24.0 | 34.0 | 0.99484 | 3.29 | 0.80 | 11.6 | 7 | 1.657143 |
| 1585 | 7.2 | 0.390 | 0.44 | 2.6 | 0.066 | 22.0 | 48.0 | 0.99494 | 3.30 | 0.84 | 11.5 | 6 | 1.916667 |
| 1586 | 7.5 | 0.310 | 0.41 | 2.4 | 0.065 | 34.0 | 60.0 | 0.99492 | 3.34 | 0.85 | 11.4 | 6 | 1.900000 |
| 1587 | 5.8 | 0.610 | 0.11 | 1.8 | 0.066 | 18.0 | 28.0 | 0.99483 | 3.55 | 0.66 | 10.9 | 6 | 1.816667 |
| 1588 | 7.2 | 0.660 | 0.33 | 2.5 | 0.068 | 34.0 | 102.0 | 0.99414 | 3.27 | 0.78 | 12.8 | 6 | 2.133333 |
| 1589 | 6.6 | 0.725 | 0.20 | 7.8 | 0.073 | 29.0 | 79.0 | 0.99770 | 3.29 | 0.54 | 9.2 | 5 | 1.840000 |
| 1590 | 6.3 | 0.550 | 0.15 | 1.8 | 0.077 | 26.0 | 35.0 | 0.99314 | 3.32 | 0.82 | 11.6 | 6 | 1.933333 |
| 1591 | 5.4 | 0.740 | 0.09 | 1.7 | 0.089 | 16.0 | 26.0 | 0.99402 | 3.67 | 0.56 | 11.6 | 6 | 1.933333 |
| 1592 | 6.3 | 0.510 | 0.13 | 2.3 | 0.076 | 29.0 | 40.0 | 0.99574 | 3.42 | 0.75 | 11.0 | 6 | 1.833333 |
| 1593 | 6.8 | 0.620 | 0.08 | 1.9 | 0.068 | 28.0 | 38.0 | 0.99651 | 3.42 | 0.82 | 9.5 | 6 | 1.583333 |
| 1594 | 6.2 | 0.600 | 0.08 | 2.0 | 0.090 | 32.0 | 44.0 | 0.99490 | 3.45 | 0.58 | 10.5 | 5 | 2.100000 |
| 1595 | 5.9 | 0.550 | 0.10 | 2.2 | 0.062 | 39.0 | 51.0 | 0.99512 | 3.52 | 0.76 | 11.2 | 6 | 1.866667 |
| 1596 | 6.3 | 0.510 | 0.13 | 2.3 | 0.076 | 29.0 | 40.0 | 0.99574 | 3.42 | 0.75 | 11.0 | 6 | 1.833333 |
| 1597 | 5.9 | 0.645 | 0.12 | 2.0 | 0.075 | 32.0 | 44.0 | 0.99547 | 3.57 | 0.71 | 10.2 | 5 | 2.040000 |
| 1598 | 6.0 | 0.310 | 0.47 | 3.6 | 0.067 | 18.0 | 42.0 | 0.99549 | 3.39 | 0.66 | 11.0 | 6 | 1.833333 |
1599 rows × 13 columns
In [20]:
#using pivot table instead of groupby to achieve same results
dframe_wine.pivot_table(index='quality')
Out[20]:
| alc / quality ratio | alcohol | chlorides | citric acid | density | fixed acidity | free sulfur dioxide | pH | residual sugar | sulphates | total sulfur dioxide | volatile acidity | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | ||||||||||||
| 3 | 3.318333 | 9.955000 | 0.122500 | 0.171000 | 0.997464 | 8.360000 | 11.000000 | 3.398000 | 2.635000 | 0.570000 | 24.900000 | 0.884500 |
| 4 | 2.566274 | 10.265094 | 0.090679 | 0.174151 | 0.996542 | 7.779245 | 12.264151 | 3.381509 | 2.694340 | 0.596415 | 36.245283 | 0.693962 |
| 5 | 1.979941 | 9.899706 | 0.092736 | 0.243686 | 0.997104 | 8.167254 | 16.983847 | 3.304949 | 2.528855 | 0.620969 | 56.513950 | 0.577041 |
| 6 | 1.771587 | 10.629519 | 0.084956 | 0.273824 | 0.996615 | 8.347179 | 15.711599 | 3.318072 | 2.477194 | 0.675329 | 40.869906 | 0.497484 |
| 7 | 1.637988 | 11.465913 | 0.076588 | 0.375176 | 0.996104 | 8.872362 | 14.045226 | 3.290754 | 2.720603 | 0.741256 | 35.020101 | 0.403920 |
| 8 | 1.511806 | 12.094444 | 0.068444 | 0.391111 | 0.995212 | 8.566667 | 13.277778 | 3.267222 | 2.577778 | 0.767778 | 33.444444 | 0.423333 |
In [22]:
dframe_wine.groupby('quality').mean()
Out[22]:
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | alc / quality ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| quality | ||||||||||||
| 3 | 8.360000 | 0.884500 | 0.171000 | 2.635000 | 0.122500 | 11.000000 | 24.900000 | 0.997464 | 3.398000 | 0.570000 | 9.955000 | 3.318333 |
| 4 | 7.779245 | 0.693962 | 0.174151 | 2.694340 | 0.090679 | 12.264151 | 36.245283 | 0.996542 | 3.381509 | 0.596415 | 10.265094 | 2.566274 |
| 5 | 8.167254 | 0.577041 | 0.243686 | 2.528855 | 0.092736 | 16.983847 | 56.513950 | 0.997104 | 3.304949 | 0.620969 | 9.899706 | 1.979941 |
| 6 | 8.347179 | 0.497484 | 0.273824 | 2.477194 | 0.084956 | 15.711599 | 40.869906 | 0.996615 | 3.318072 | 0.675329 | 10.629519 | 1.771587 |
| 7 | 8.872362 | 0.403920 | 0.375176 | 2.720603 | 0.076588 | 14.045226 | 35.020101 | 0.996104 | 3.290754 | 0.741256 | 11.465913 | 1.637988 |
| 8 | 8.566667 | 0.423333 | 0.391111 | 2.577778 | 0.068444 | 13.277778 | 33.444444 | 0.995212 | 3.267222 | 0.767778 | 12.094444 | 1.511806 |
In [24]:
#lets plot the data on a scatterplot
%matplotlib inline
dframe_wine.plot(kind='scatter', x='quality', y='alcohol')
Out[24]:
<matplotlib.axes._subplots.AxesSubplot at 0x1a71c4c93c8>